We Are Problem Solvers Driven By Outcomes
SARS-CoV-2 Life Cycle and Opportunities
for Intervention
A recent S-protein mutation (November 2019) is believed to have enabled the virus to infect humans.1
Intervention OpportunitiesThere are 3 main approaches to preventing and treating COVID-19:
- Prevent viral entry into the host cell
- Prevent viral replication
- Prevent viral actions and reactions of the host
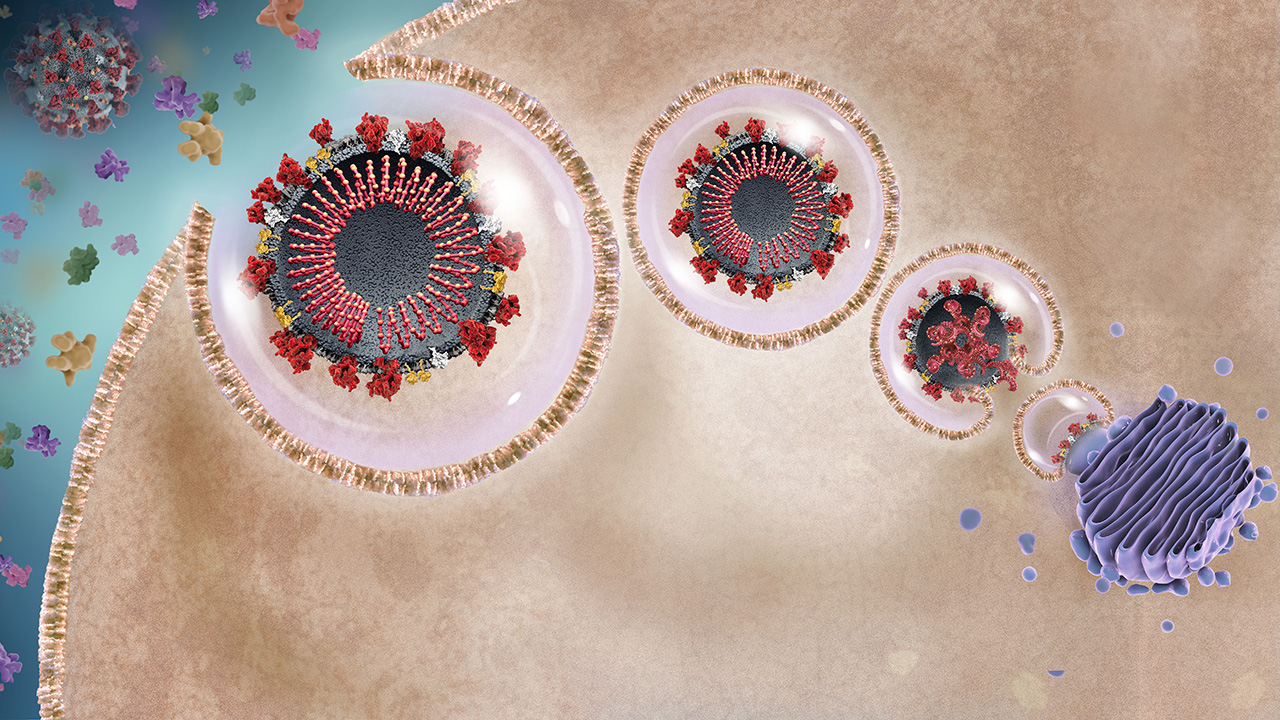
SARS-CoV-2 and Host Cell Interaction
The host cell receptor ACE2 binds the viral spike protein to enable viral entry into the cell.1-3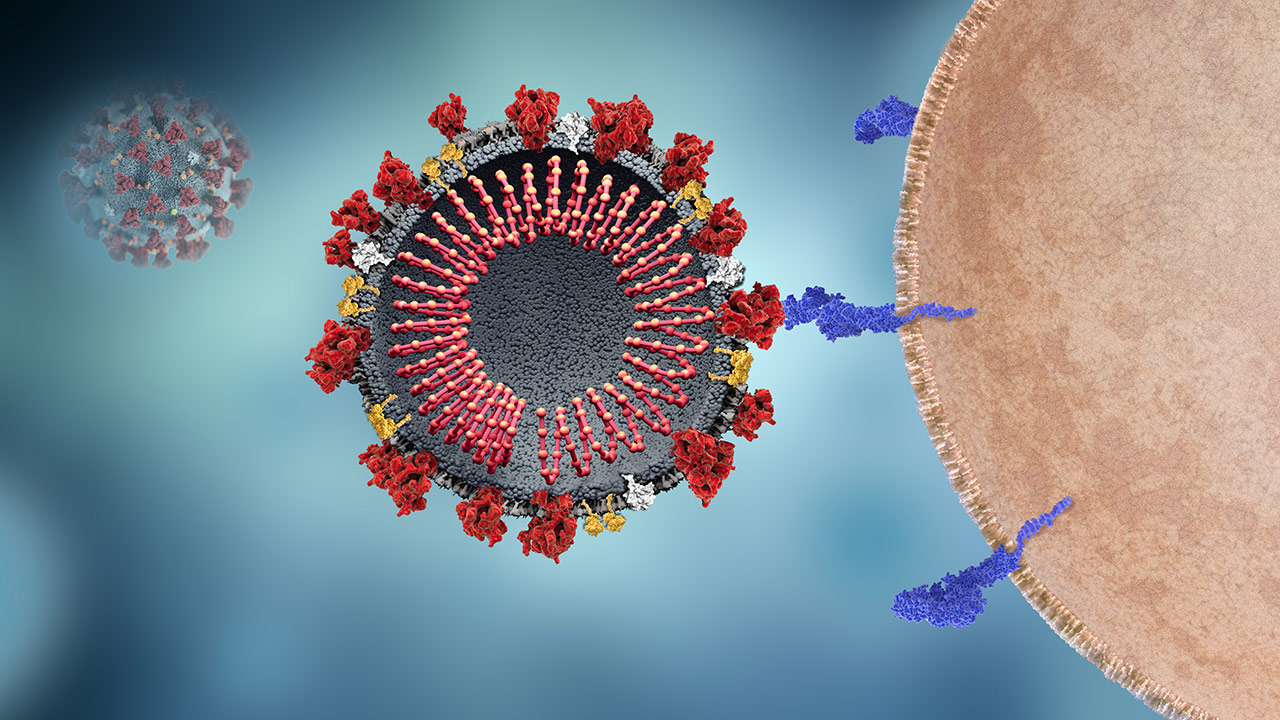
to learn more.
Severe acute respiratory syndrome–associated coronavirus 2 (SARS-CoV-2)
OverviewSARS-CoV-2 is a positive-stranded RNA virus named for its crownlike microscopic appearance. SARS-CoV-2 causes COVID-19 (CoronaVirus Disease-19).1
Membrane (M) protein
OverviewThe M protein, the most abundant of the 4 critical structural proteins, gives the virus its shape. It is critical in directing assembly of the daughter viruses for release.2,3
Intervention OpportunitiesThe M protein is critical to viral assembly and structure; interfering with its actions could be an opportunity to target the virus.11
Envelope (E) protein
OverviewThe E protein, an envelope transmembrane structural protein, assists the M protein in directing assembly of the daughter viruses.2,3
Intervention OpportunitiesIn other coronaviruses, deletion of the E protein decreases the virus’s ability to replicate. Live attenuated viruses with E protein deletions may be vaccine candidates.10
Nucleocapsid (N) protein
OverviewThe N protein, 1 of the 4 critical structural proteins, binds the viral RNA creating the nucleocapsid. It is critical to supporting assembly of the daughter viruses for release.2,3
Intervention OpportunitiesThe N protein has been identified as a potential target for vaccines.5
RNA
OverviewThe coronavirus contains a single strand of positive RNA, which directs the production of the necessary proteins to create new infective viruses with minimal host cell machinery in the cell cytoplasm.3
Intervention OpportunitiesMany opportunities exist to inhibit the RNA from reaching the host cell machinery and inhibiting its ability to direct its own replication.12-15
Spike (S) protein
OverviewThe S protein, 1 of 4 critical structural proteins, interacts with receptor ACE2 of the host cell membrane to enable viral attachment and entrance into the host cell.2,3,4 It appears that a recent S-protein mutation (November 2019) allowed this virus to infect humans.1
Intervention OpportunitiesThe S2 subunit of the S protein is highly conserved, which makes it a potential target for antiviral compounds and vaccines.1,3,5 For example, monoclonal antibodies to it may inhibit its action.4
Receptor ACE2
OverviewThe virus binds the host cell receptor ACE2 protein, a critical step in viral entry.3
Intervention OpportunitiesACE2 is part of the renin-angiotensin system (RAS). The effects of the virus on the RAS are not clearly understood.5 It is unclear whether the effects of RAS drugs commonly used for blood pressure control are beneficial or harmful in COVID-19.6-8 Trials are ongoing investigating ACE2, ACE, and products of the RAS.7-9
Envelope
OverviewSARS-CoV-2 is an enveloped virus. The envelope is a phospholipid wrapping derived from the host cell membrane. The envelope promotes viral assembly and release and is thus critical in virus pathogenicity.1
Intervention OpportunitiesThe envelope is made up of phospholipids and is sensitive to destruction by alcohol, heat, UV light, or soap.1
SARS-CoV-2 Enters Host Cell
The coronavirus enters the cell, encapsulated by the host cell’s membrane.3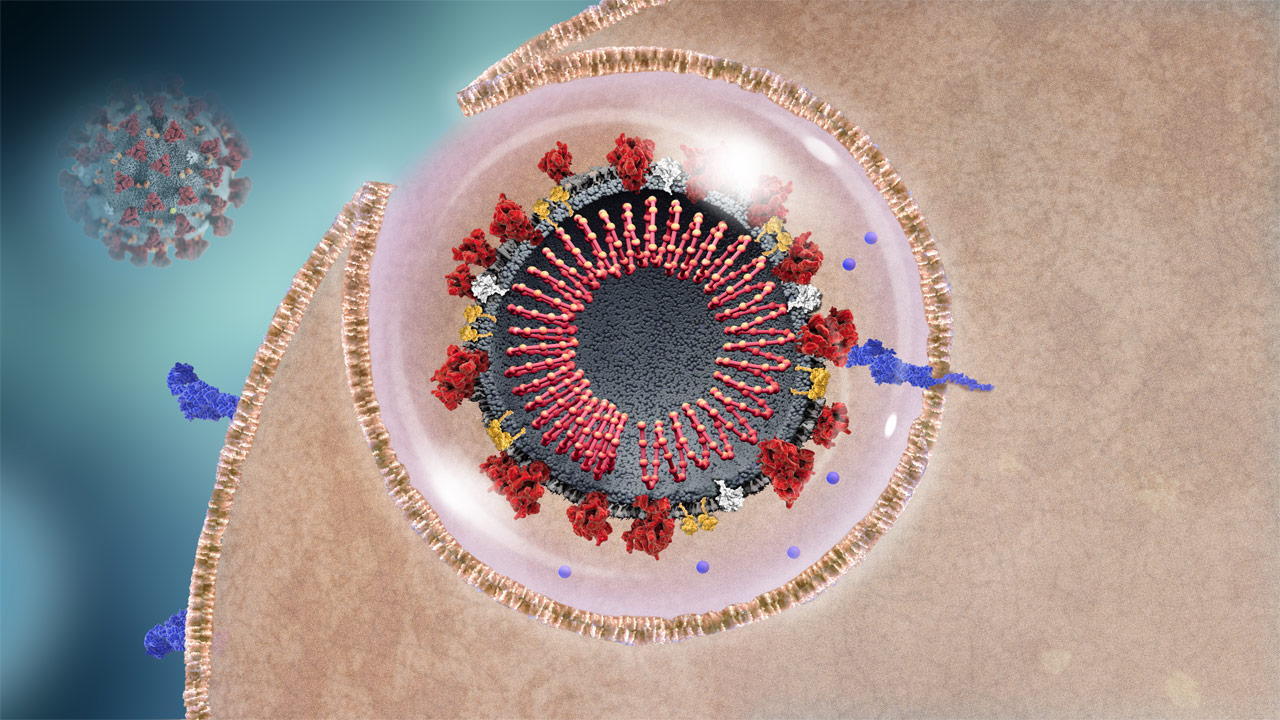
to learn more.
Virus entry
OverviewThe virus enters the host cell through endocytosis, encapsulated in a piece of the host cell membrane.3,14
Intervention OpportunitiesVarious drugs are proposed to affect and inhibit the viral entrance into the host cell; however, the specific mechanisms have not been elucidated. For example, macrolide antibiotics may inhibit viral entry.12
Viral RNA Release Into Host Cell
The virus initially enters the cell in an endosome. The endosome fuses with the viral envelope to release the viral RNA into the cell cytoplasm.2,3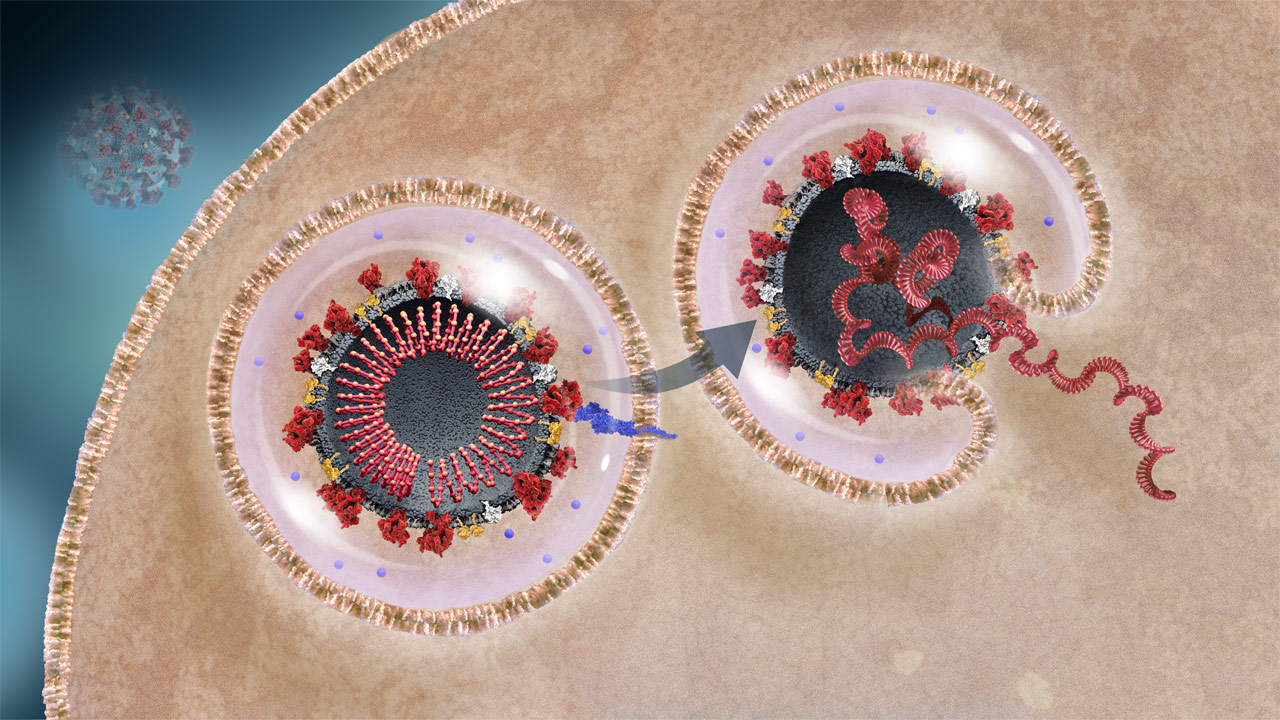
to learn more.
Endosome
OverviewThe virus enters the cell encapsulated in a piece of the host cell membrane, an endosome, from which release is necessary for replication.3
Intervention OpportunitiesA slightly acidic pH in the endosome is critical for RNA release.10 Antimalarial drugs may increase the pH within the endosome and prevent release of the RNA and thus subsequent viral activity.13
TMPRRS2
OverviewTMPRRS2 is a host cell protease that cleaves the S protein, enabling fusion of the viral and cellular membranes, releasing viral RNA into the host cell. An acidic environment is necessary for this enzyme activity.3
Intervention OpportunitiesTMPRRS2 is one of numerous proteases necessary for viral reproduction. It needs a slightly acidic environment to function.3 Antimalarials may increase the pH in the endosome.13
RNA
OverviewThe coronavirus contains a single strand of positive RNA, which directs the production of the necessary proteins to create new infective viruses with minimal host cell machinery in the cell cytoplasm.3
Intervention OpportunitiesMany opportunities exist to inhibit the RNA from reaching the host cell machinery and inhibiting its ability to direct its own replication.12-15
Replicase-Transcriptase Complex Production
Various nsps are produced through RNA transcription and polypeptide cleavage. Replicase is translated from the RNA and processed to a functional enzyme. The replicase-transcriptase complex (RTC) will include the viral RNA, replicase enzyme, and nsps.2,3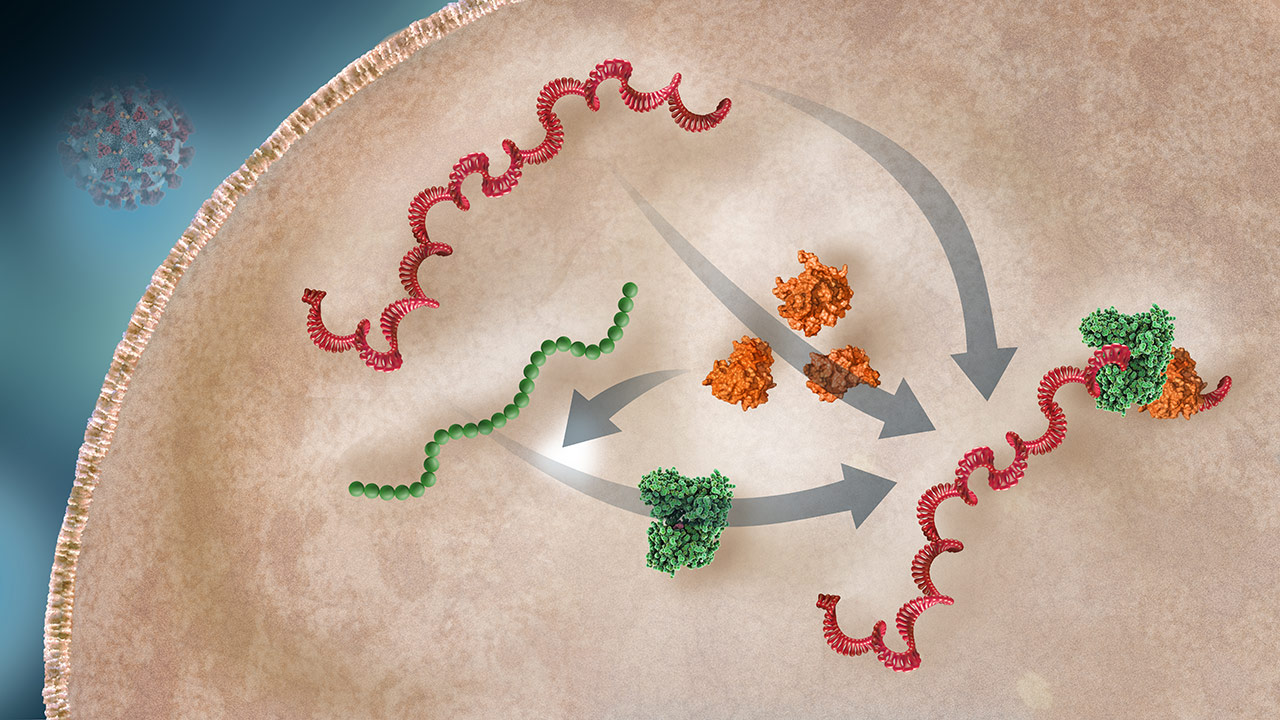
to learn more.
RNA
OverviewThe coronavirus contains a single strand of positive RNA, which directs the production of the necessary proteins to create new infective viruses with minimal host cell machinery in the cell cytoplasm.3
Intervention OpportunitiesMany opportunities exist to inhibit the RNA from reaching the host cell machinery and inhibiting its ability to direct its own replication.12-15
Replicase
OverviewReplicase is a critical enzyme that transcribes (duplicates) the RNA, creating RNA for daughter viruses and mRNA for viral protein translation (production).3
Intervention OpportunitiesReplicase inhibitors are under investigation for treatment and prevention of COVID-19. One lead compound is remdesivir, which substitutes adenosine analogues and prevents proper translation.18
Nonstructural Proteins (nsps)
OverviewVarious nsps support the viral replication process. Papain-like protease (PLpro) cleaves a polypeptide precursor of replicase.3 Other nsps are integral parts of the replicase-transcriptase complex (RTC).1,3 Of note, some nsps block the host innate immune response.1
Intervention OpportunitiesProteases are theoretically potential targets for protease inhibitors.11 Protease inhibitors have been used in other viral diseases such as HIV and may have activity against SARS-CoV-2.16,17
Replicase-Transcriptase Complex (RTC)
OverviewThe RTC transcribes (duplicates) the RNA, creating RNA for daughter viruses and mRNA for viral protein translation (production).3
Intervention OpportunitiesReplicase inhibitors are under investigation for treatment and prevention of COVID-19. One lead compound is remdesivir, which substitutes adenosine analogues and prevents proper translation.18
Viral RNA Transcription
The viral RNA is used as the map to transcribe both mRNA (which will be used to produce viral proteins) and full copies of the RNA (for daughter virus production).2,3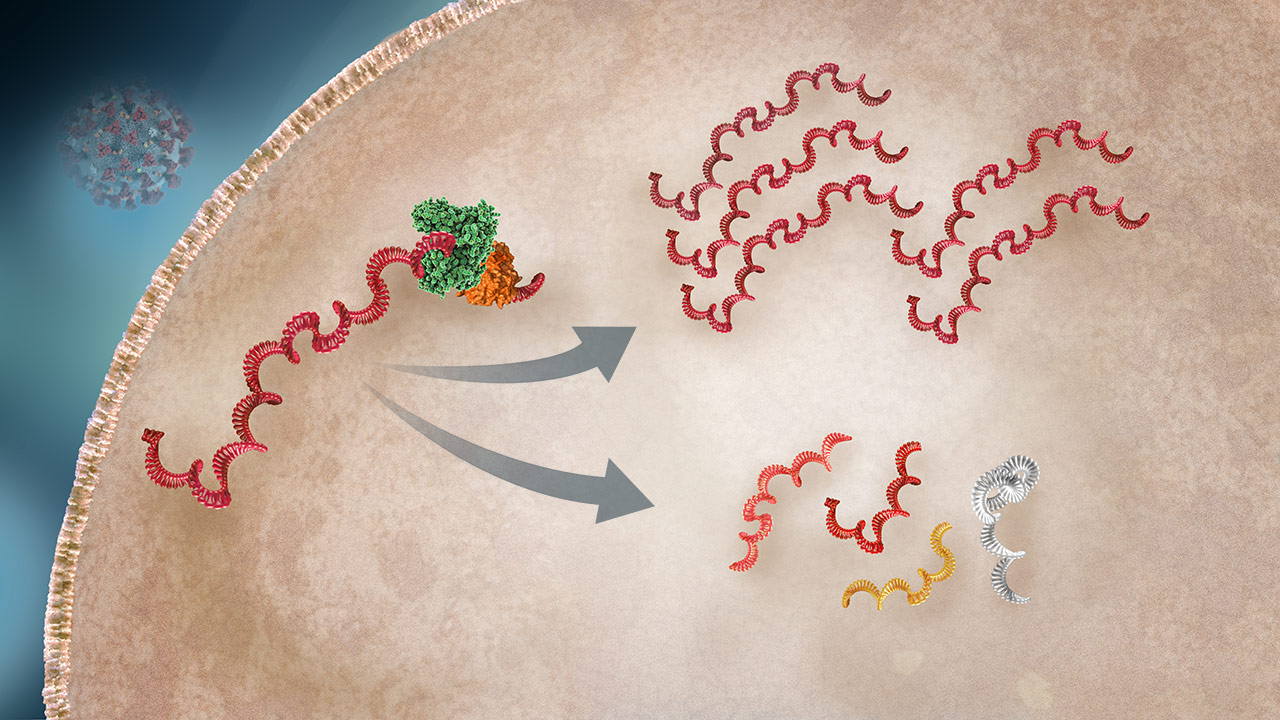
to learn more.
Replicase-transcriptase Complex
OverviewThe RTC transcribes (duplicates) the RNA, creating RNA for daughter viruses and mRNA for viral protein translation (production).3
Intervention OpportunitiesReplicase inhibitors are under investigation for treatment and prevention of COVID-19. One lead compound is remdesivir, which substitutes adenosine analogues and prevents proper translation.18
Genomic RNA
OverviewComplete genomic RNA transcripts are the single strands of RNA that will be incorporated into new viruses.3
Intervention OpportunitiesVarious opportunities exist to prevent the incorporation of the new transcripts, including inhibiting replication. One lead compound is remdesivir, which substitutes adenosine analogues and prevents proper translation.18
Subgenomic (sg) RNA
OverviewVarious sgRNA transcripts serve as the mRNA to guide production of proteins to be incorporated into new viruses.3
Intervention OpportunitiesPrevention of mRNA translation into new proteins could prevent viral formation. RNA interference (RNAi) has been studied in other coronaviruses.19
Protein Production
The host cell’s machinery translates mRNA into viral proteins.2,3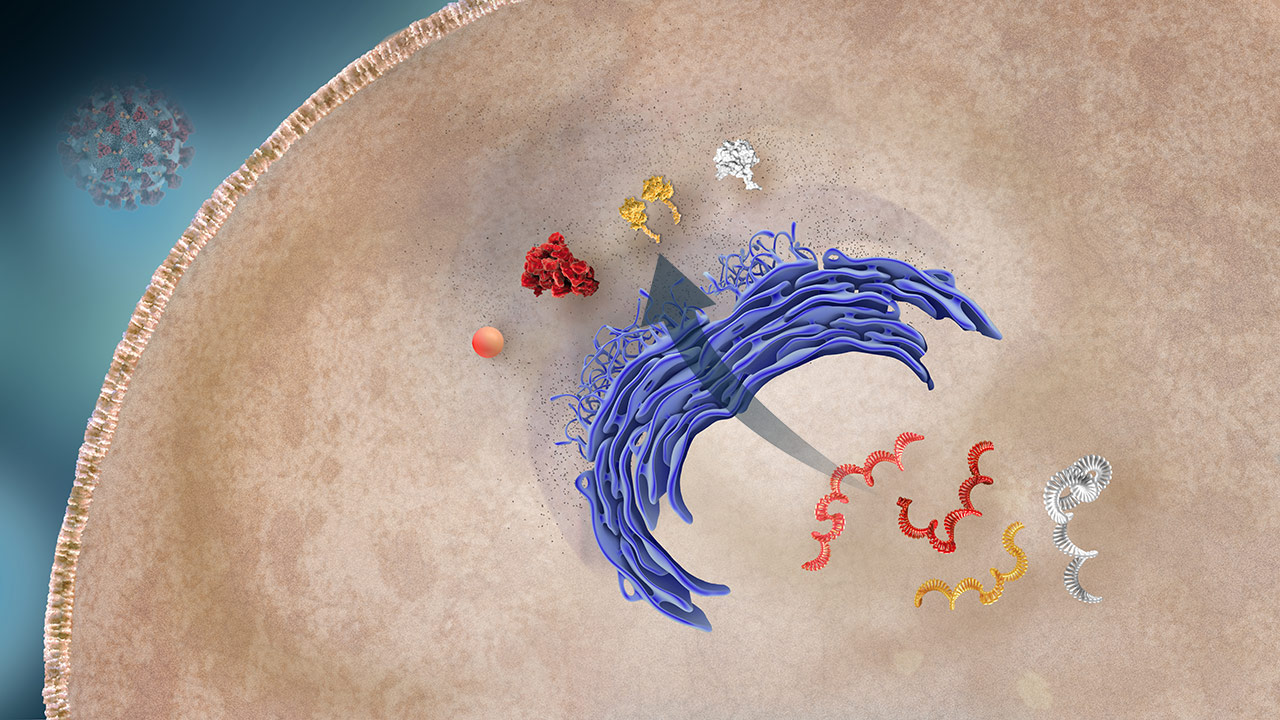
to learn more.
Structural Proteins
OverviewThere are 4 main structural proteins: S, M, E, and N. The viral mRNA directs the host cell machinery (endoplasmic reticulum and golgi apparatus) to produce and package them into the daughter viruses.3
Intervention OpportunitiesEach of these proteins serves specific viral functions; interfering with or inhibiting them could inhibit the virus. Furthermore, these may be targets for vaccines.5,15
Subgenomic (sg) RNA
OverviewVarious sgRNA transcripts serve as the mRNA to guide production of proteins to be incorporated into new viruses.3
Intervention OpportunitiesPrevention of mRNA translation into new proteins could prevent viral formation. RNA interference (RNAi) has been studied in other coronaviruses.19
Protein Packaging and Organization Into Daughter Viruses
The N protein organizes the RNA, creating the nucleocapsid. The various envelope constituent viral proteins are packaged into the lipid bilayer (the future viral envelopes), which encapsulates the nucleocapsid.2,3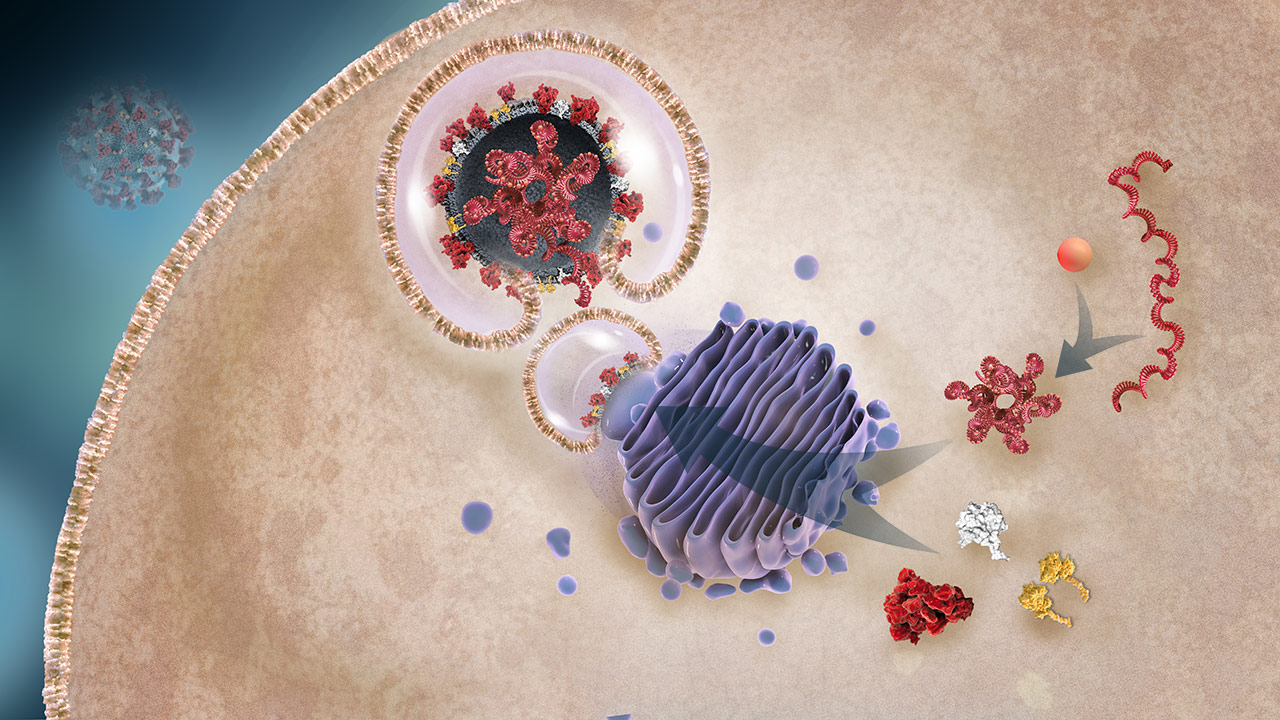
to learn more.
RNA
OverviewThe coronavirus contains a single strand of positive RNA, which directs the production of the necessary proteins to create new infective viruses with minimal host cell machinery in the cell cytoplasm.3
Intervention OpportunitiesMany opportunities exist to inhibit the RNA from reaching the host cell machinery and inhibiting its ability to direct its own replication.12-15
Nucleocapsid (N) protein
OverviewThe N protein, 1 of the 4 critical structural proteins, binds the viral RNA creating the nucleocapsid. It is critical to supporting assembly of the daughter viruses for release.2,3
Intervention OpportunitiesThe N protein has been identified as a potential target for vaccines.5
Nucleocapsid
OverviewN protein binds RNA and organizes it, forming the nucleocapsid.2
Intervention OpportunitiesPrevention of nucleocapsid formation could be a target to inhibit viral formation.18 The N protein is a major constituent of the nucleocapsid and a potential target for vaccines.5
Envelope (E) protein
OverviewThe E protein, an envelope transmembrane structural protein, assists the M protein in directing assembly of the daughter viruses.2,3
Intervention OpportunitiesIn other coronaviruses, deletion of the E protein decreases the virus’s ability to replicate. Live attenuated viruses with E protein deletions may be vaccine candidates.10
Membrane (M) protein
OverviewThe M protein, the most abundant of the 4 critical structural proteins, gives the virus its shape. It is critical in directing assembly of the daughter viruses for release.2,3
Intervention OpportunitiesThe M protein is critical to viral assembly and structure; interfering with its actions could be an opportunity to target the virus.11
Spike (S) protein
OverviewThe S protein, 1 of 4 critical structural proteins, interacts with receptor ACE2 of the host cell membrane to enable viral attachment and entrance into the host cell.2,3,4 It appears that a recent S-protein mutation (November 2019) allowed this virus to infect humans.1
Intervention OpportunitiesThe S2 subunit of the S protein is highly conserved, which makes it a potential target for antiviral compounds and vaccines.1,3,5 For example, monoclonal antibodies to it may inhibit its action.4
New Virus Release
The endosome fuses with the cell membrane and releases the active daughter viruses, which can infect other cells within the host or other people.2 Throughout infection, the host produces a variety of chemicals and cytokines called a cytokine storm.20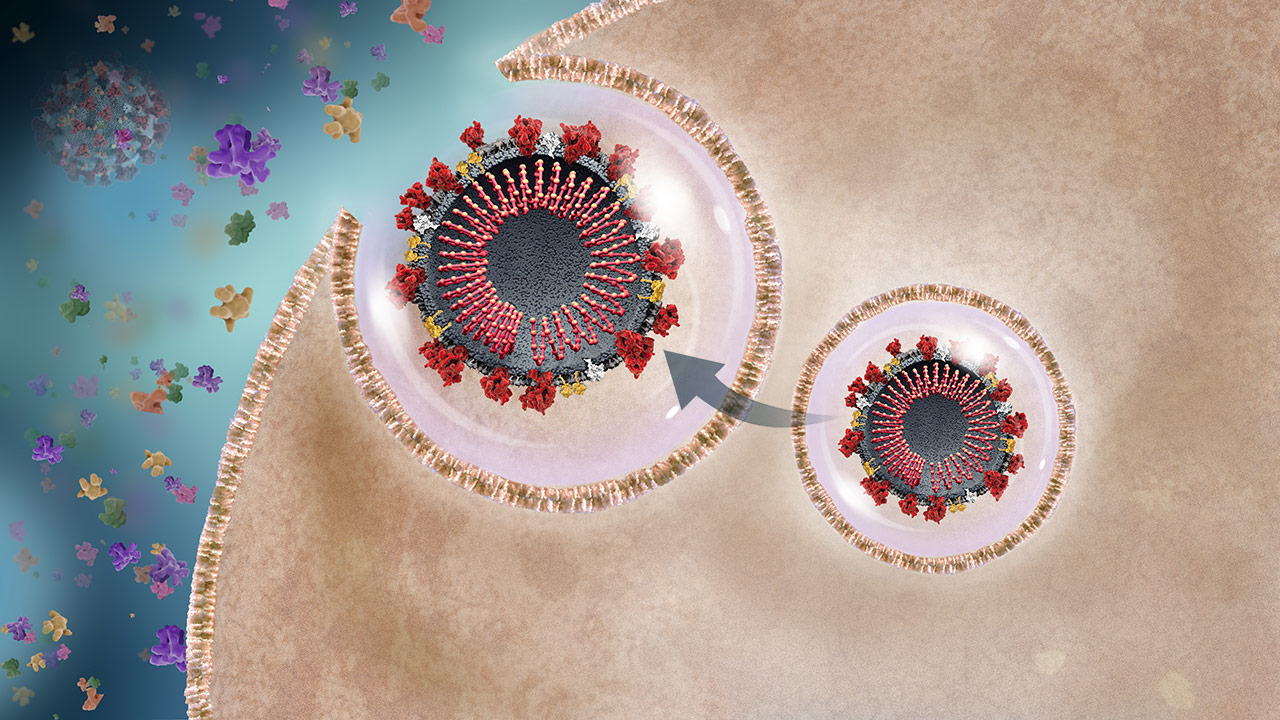
to learn more.
Severe acute respiratory syndrome–associated coronavirus 2 (SARS-CoV-2)
OverviewSARS-CoV-2 is a positive-stranded RNA virus named for its crownlike microscopic appearance. SARS-CoV-2 causes COVID-19 (CoronaVirus Disease-19).1
Cytokine Storm
OverviewThroughout infection, the host produces a variety of chemicals and cytokines called a cytokine storm. These are implicated in the immune response and observed morbidity and mortality of COVID-19.20
Intervention OpportunitiesWhile inhibition of cytokines may not affect the viral life cycle, it may reduce overall symptoms, morbidity, and mortality. Various cytokines and their immune responses are targets for therapy, including interleukin (IL)-6 inhibitors, Janus kinase (JAK) inhibitors, interferons, and others.9,20
References
- Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation and treatment coronavirus (COVID-19). StatPearls – NCBI Bookshelf. 2020;1:1-17.
- Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16(1):16-69.
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In: Maier HJ, et al, eds. Coronaviruses: Methods and Protocols, Methods in Molecular Biology. Vol. 1282. New York, NY: Springer Science+Business Media; 2015:1-23.
- Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspective [published online ahead of print]. Int J Antimicrob Agents. 2020. doi:10.1016/j.ijantimicag.2020.105951.
- Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):1-15.
- Bao H, Gao F, Xie G, Liu Z. Angiotensin-converting enzyme 2 inhibits apoptosis of pulmonary endothelial cells during acute lung injury through suppressing MiR-4262. Cell Physiol Biochem. 2015;37:759-767.
- Malha L, Mueller FB, Pecker MS, Mann SJ, August P, Feig PU. COVID-19 and the renin-angiotensin system. Kidney Int Rep. 2020:1-3. doi:10.1016/j.ekir.2020.03.024. In press.
- NIH US National Library of Medicine. Recombinant human angiotensin-converting enzyme 2 (rhACE2) as a treatment for patients with COVID-19 (APN01-COVID-19). Clinical Trials.gov. https://clinicaltrials.gov/ct2/show/NCT04335136. Accessed April 16, 2020.
- US National Library of Medicine. Clinicaltrials.gov. Accessed April 22, 2020.
- Netland J, DeDiego ML, Zhao J, et al. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology. 2010;399(1):120-128.
- Prajapa M, Sarma P, Shekhar N, et al. Drug targets for corona virus: a systematic review. Indian J Pharmacol. 2020:56-65.
- Tran DH, Sugamata R, Hirose T, et al. Azithromycin, a 15-membered macrolide antibiotic, inhibits influenza A (H1N1) pdm09 virus infection by interfering with virus internalization process. J Antibiot (Tokyo). 2019;72(10):759-768.
- Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2(1):69.
- Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci. 2020;16(10):1724-1731.
- Timmer J. COVID-19: the biology of an effective therapy. ARS Technica website. https://arstechnica.com/science/2020/03/covid-19-the-biology-of-an-effective-therapy/. Accessed April 22, 2020.
- Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov Ther. 2020;14(1):58-60.
- Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59(14):6595-6628.
- Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus [published online ahead of print February 24, 2020]. J Biol Chem. 2020. doi:10.1074/jbc.AC120.013056.
- Zhang Y, Li T, Fu L, Yu C, Li Y, Xu X. Silencing SARS‐CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560:141-146.
- Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033-1034.
